7 易生信-数据可视化案例分享
从一套表达和通路数据入手,探索常见的绘图展示方式和报错处理。
7.1 加载需要的包
7.2 读入数据
7.2.1 ’row.names’里不能有重复的名字 Duplicate row names
7.2.2 行名唯一化处理
这里使用make.names转换行名为唯一,实际需要先弄清楚为什么会有重复名字。
expr <- read.table("ehbio.simplier.DESeq2.normalized.symbol.txt", row.names = NULL, header = T, sep = "\t")
head(expr)## id untrt_N61311 untrt_N052611 untrt_N080611 untrt_N061011 trt_N61311
## 1 FN1 245667.66 427435.1 221687.51 371144.2 240187.24
## 2 DCN 212953.14 360796.2 258977.30 408573.1 210002.18
## 3 CEMIP 40996.34 137783.1 53813.92 91066.8 62301.12
## 4 CCDC80 137229.15 232772.2 86258.13 212237.3 136730.76
## 5 IGFBP5 77812.65 288609.2 210628.87 168067.4 96021.74
## 6 COL1A1 146450.41 127367.3 152281.50 140861.1 62358.64
## trt_N052611 trt_N080611 trt_N061011
## 1 450103.21 280226.19 376518.23
## 2 316009.14 225547.39 393843.74
## 3 223111.85 212724.84 157919.47
## 4 226070.89 124634.56 236237.81
## 5 217439.21 162677.38 168387.36
## 6 53800.47 69160.97 51044.06有哪些基因名是重复出现的?
## [1] "MATR3" "PKD1P1" "HSPA14" "OR7E47P" "POLR2J3"
## [6] "ATXN7" "TMSB15B" "LINC-PINT" "TBCE" "SNX29P2"
## [11] "SCO2" "POLR2J4" "CCDC39" "RGS5" "BMS1P21"
## [16] "RF00017" "GOLGA8M" "RF00017" "DNAJC9-AS1" "CYB561D2"
## [21] "RF00017" "IPO5P1" "RF00017" "RF00017" "RF00017"
## [26] "SPATA13" "RF00017" "RF00017" "RF00017" "RF00017"
## [31] "RF00017" "RF00017" "RF00017" "RF00017" "RF00017"
## [36] "RF00017" "RF00017" "RF00017" "RF00017" "RF00017"
## [41] "RF00019" "RF00019" "RF00017" "RF00017" "RF00017"
## [46] "RF00019" "BMS1P4" "RF00019" "RF00019" "RF00017"
## [51] "RF00017" "RF00017" "RF00017" "RF00017" "RF00017"
## [56] "RF00017" "RF00017" "RF00017" "RF00017" "RF00017"
## [61] "RF00017" "RF00017" "RF00017" "RF00019" "RF00017"
## [66] "RF00017" "RF00017" "RF00019" "RF00017" "RF00017"
## [71] "LINC01238" "RF00017" "RF00017" "RF00017" "RF00017"
## [76] "RF00017" "RF00017" "RF00017" "RF00017" "RF00017"
## [81] "RF00017" "RF00017" "RF02271" "RF00017" "RF00017"
## [86] "RF00017" "RF00017" "RF00017" "LINC01297" "RF00019"
## [91] "RF00017" "RF00012" "RF00019" "RF00017" "RF00017"
## [96] "RF00019" "RF00017" "RF00017" "RF00017" "ZNF503"
## [101] "RF00017" "RF00017" "RF00017" "RF00017" "RF00017"
## [106] "RF00017" "RF00017" "RF00017" "RF02271" "RF00019"
## [111] "RF00019" "RF00017" "RF00019" "RF02271" "RF00017"
## [116] "RF00017" "RF00017" "RF00017" "RF00019" "RF00019"
## [121] "RF00017" "RF00019" "ITFG2-AS1" "RF00019" "RF00019"
## [126] "RF00017" "RF00019" "RF00017" "RF00017" "RF00017"
## [131] "RF00019" "RF00017" "RF00012" "RF00017" "RF00017"
## [136] "RAET1E-AS1" "RF00017" "RF00017" "RF00017" "RF00017"
## [141] "RF00017" "RF00017" "RF00017" "RF00017" "RF00012"
## [146] "RF02271" "RF00019" "LINC01422" "RF02271" "RF00017"
## [151] "RF00019" "RF00019" "RF00019" "RF00019" "RF00017"
## [156] "LINC01481" "RF00017" "SNHG28" "RF00019" "RF00019"
## [161] "RF00019" "RF00019" "LINC00484" "LINC00941" "ALG1L9P"
## [166] "RF00017" "DUXAP8" "RF00017" "RF00017" "RF00017"
## [171] "RF00017" "RF00017" "RF00017" "RMRP" "RF00017"
## [176] "RF00017" "RF00017" "RF00017" "DIABLO"名字唯一化处理
## [1] "a" "a.1" "b" "b.1" "b.2"唯一化之后的名字作为行名字,并去掉原来的第一列
expr_names <- make.names(expr$id, unique = T)
rownames(expr) <- expr_names
expr <- expr[, -1]
head(expr)## untrt_N61311 untrt_N052611 untrt_N080611 untrt_N061011 trt_N61311
## FN1 245667.66 427435.1 221687.51 371144.2 240187.24
## DCN 212953.14 360796.2 258977.30 408573.1 210002.18
## CEMIP 40996.34 137783.1 53813.92 91066.8 62301.12
## CCDC80 137229.15 232772.2 86258.13 212237.3 136730.76
## IGFBP5 77812.65 288609.2 210628.87 168067.4 96021.74
## COL1A1 146450.41 127367.3 152281.50 140861.1 62358.64
## trt_N052611 trt_N080611 trt_N061011
## FN1 450103.21 280226.19 376518.23
## DCN 316009.14 225547.39 393843.74
## CEMIP 223111.85 212724.84 157919.47
## CCDC80 226070.89 124634.56 236237.81
## IGFBP5 217439.21 162677.38 168387.36
## COL1A1 53800.47 69160.97 51044.067.3 热图绘制
 ### 提取差异基因绘制热图 {#visual6}
### 提取差异基因绘制热图 {#visual6}
读入差异基因列表
de_gene <- read.table("ehbio.DESeq2.all.DE.symbol", row.names = NULL, header = F, sep = "\t")
head(de_gene)## V1 V2
## 1 ARHGEF2 untrt._higherThan_.trt
## 2 KCTD12 untrt._higherThan_.trt
## 3 SLC6A9 untrt._higherThan_.trt
## 4 GXYLT2 untrt._higherThan_.trt
## 5 RAB7B untrt._higherThan_.trt
## 6 NEK10 untrt._higherThan_.trt提取Top3 差异的基因
# library(dplyr)
top6_de_gene <- de_gene %>% group_by(V2) %>% dplyr::slice(1:3)
top6 <- expr[which(rownames(expr) %in% top6_de_gene$V1), ]
head(top6)## untrt_N61311 untrt_N052611 untrt_N080611 untrt_N061011 trt_N61311
## KCTD12 4700.79369 3978.0401 4416.15169 4792.34174 936.69481
## MAOA 438.54451 452.9934 516.63033 258.73279 4628.00860
## ARHGEF2 3025.62334 3105.7830 3094.51304 2909.99043 1395.39850
## SPARCL1 58.15705 102.5827 80.00997 82.59042 2220.50867
## PER1 170.61639 156.3692 194.97497 123.47689 1728.38117
## SLC6A9 360.66314 413.8797 365.47650 443.71982 63.90538
## trt_N052611 trt_N080611 trt_N061011
## KCTD12 633.4462 979.77576 641.49582
## MAOA 4429.7201 4629.66529 3778.17351
## ARHGEF2 1441.9916 1464.59769 1501.51509
## SPARCL1 1750.9879 1374.90745 2194.58930
## PER1 1230.2575 1120.00650 1333.91208
## SLC6A9 56.8962 86.82929 95.33916读入样品分组信息作为列注释
metadata <- read.table("sampleFile", header = T, row.names = 1)
pheatmap(top6, annotation_col = metadata)
按行标准化后展示

7.4 箱线图和统计比较
## untrt_N61311 untrt_N052611 untrt_N080611 untrt_N061011 trt_N61311
## KCTD12 4700.79369 3978.0401 4416.15169 4792.34174 936.69481
## MAOA 438.54451 452.9934 516.63033 258.73279 4628.00860
## ARHGEF2 3025.62334 3105.7830 3094.51304 2909.99043 1395.39850
## SPARCL1 58.15705 102.5827 80.00997 82.59042 2220.50867
## PER1 170.61639 156.3692 194.97497 123.47689 1728.38117
## SLC6A9 360.66314 413.8797 365.47650 443.71982 63.90538
## trt_N052611 trt_N080611 trt_N061011
## KCTD12 633.4462 979.77576 641.49582
## MAOA 4429.7201 4629.66529 3778.17351
## ARHGEF2 1441.9916 1464.59769 1501.51509
## SPARCL1 1750.9879 1374.90745 2194.58930
## PER1 1230.2575 1120.00650 1333.91208
## SLC6A9 56.8962 86.82929 95.33916矩阵转置
## KCTD12 MAOA ARHGEF2 SPARCL1 PER1 SLC6A9
## untrt_N61311 4700.7937 438.5445 3025.623 58.15705 170.6164 360.66314
## untrt_N052611 3978.0401 452.9934 3105.783 102.58269 156.3692 413.87971
## untrt_N080611 4416.1517 516.6303 3094.513 80.00997 194.9750 365.47650
## untrt_N061011 4792.3417 258.7328 2909.990 82.59042 123.4769 443.71982
## trt_N61311 936.6948 4628.0086 1395.398 2220.50867 1728.3812 63.90538
## trt_N052611 633.4462 4429.7201 1441.992 1750.98786 1230.2575 56.89620
## trt_N080611 979.7758 4629.6653 1464.598 1374.90745 1120.0065 86.82929
## trt_N061011 641.4958 3778.1735 1501.515 2194.58930 1333.9121 95.33916与样本属性信息合并
## Row.names conditions individual KCTD12 MAOA ARHGEF2 SPARCL1
## 1 trt_N052611 trt N052611 633.4462 4429.7201 1441.992 1750.98786
## 2 trt_N061011 trt N061011 641.4958 3778.1735 1501.515 2194.58930
## 3 trt_N080611 trt N080611 979.7758 4629.6653 1464.598 1374.90745
## 4 trt_N61311 trt N61311 936.6948 4628.0086 1395.398 2220.50867
## 5 untrt_N052611 untrt N052611 3978.0401 452.9934 3105.783 102.58269
## 6 untrt_N061011 untrt N061011 4792.3417 258.7328 2909.990 82.59042
## PER1 SLC6A9
## 1 1230.2575 56.89620
## 2 1333.9121 95.33916
## 3 1120.0065 86.82929
## 4 1728.3812 63.90538
## 5 156.3692 413.87971
## 6 123.4769 443.71982修改第一列的列名字
## Sample conditions individual KCTD12 MAOA ARHGEF2 SPARCL1
## 1 trt_N052611 trt N052611 633.4462 4429.7201 1441.992 1750.98786
## 2 trt_N061011 trt N061011 641.4958 3778.1735 1501.515 2194.58930
## 3 trt_N080611 trt N080611 979.7758 4629.6653 1464.598 1374.90745
## 4 trt_N61311 trt N61311 936.6948 4628.0086 1395.398 2220.50867
## 5 untrt_N052611 untrt N052611 3978.0401 452.9934 3105.783 102.58269
## 6 untrt_N061011 untrt N061011 4792.3417 258.7328 2909.990 82.59042
## PER1 SLC6A9
## 1 1230.2575 56.89620
## 2 1333.9121 95.33916
## 3 1120.0065 86.82929
## 4 1728.3812 63.90538
## 5 156.3692 413.87971
## 6 123.4769 443.719827.4.1 单基因箱线图
library(ggpubr)
ggboxplot(top6_t_with_group, x = "conditions", y = "KCTD12", title = "KCTD12", ylab = "Expression", color = "conditions",
palette = "jco")
# palette npg, lancet,
7.4.2 多基因箱线图 (combine)
ggboxplot(top6_t_with_group, x = "conditions", y = c("KCTD12", "MAOA", "PER1", "SLC6A9"), ylab = "Expression", combine = T,
color = "conditions", palette = "jco")
7.4.3 多基因箱线图 (merge)
ggboxplot(top6_t_with_group, x = "conditions", y = c("KCTD12", "MAOA", "PER1", "SLC6A9"), ylab = "Expression", merge = "flip",
color = "conditions", palette = "nature")
7.4.4 数据对数转换后绘制箱线图
top6_t_with_group_log = top6_t_with_group %>% purrr::map_if(is.numeric, log1p) %>% as.data.frame
head(top6_t_with_group_log)## Sample conditions individual KCTD12 MAOA ARHGEF2 SPARCL1
## 1 trt_N052611 trt N052611 6.452752 8.396317 7.274474 7.468506
## 2 trt_N061011 trt N061011 6.465360 8.237261 7.314896 7.694206
## 3 trt_N080611 trt N080611 6.888344 8.440456 7.290018 7.226869
## 4 trt_N61311 trt N61311 6.843425 8.440098 7.241652 7.705942
## 5 untrt_N052611 untrt N052611 8.288796 6.118083 8.041343 4.640370
## 6 untrt_N061011 untrt N061011 8.474983 5.559653 7.976249 4.425929
## PER1 SLC6A9
## 1 7.115791 4.058652
## 2 7.196621 4.567875
## 3 7.021982 4.475395
## 4 7.455519 4.172930
## 5 5.058595 6.027989
## 6 4.824120 6.097444ggboxplot(top6_t_with_group_log, x = "conditions", y = c("KCTD12", "MAOA", "PER1", "SLC6A9"), ylab = "Expression", merge = "flip",
fill = "conditions", palette = "Set3")
7.4.5 用ggplot2实现ggpubr
## Sample conditions individual KCTD12 MAOA ARHGEF2 SPARCL1
## 1 trt_N052611 trt N052611 633.4462 4429.7201 1441.992 1750.98786
## 2 trt_N061011 trt N061011 641.4958 3778.1735 1501.515 2194.58930
## 3 trt_N080611 trt N080611 979.7758 4629.6653 1464.598 1374.90745
## 4 trt_N61311 trt N61311 936.6948 4628.0086 1395.398 2220.50867
## 5 untrt_N052611 untrt N052611 3978.0401 452.9934 3105.783 102.58269
## 6 untrt_N061011 untrt N061011 4792.3417 258.7328 2909.990 82.59042
## PER1 SLC6A9
## 1 1230.2575 56.89620
## 2 1333.9121 95.33916
## 3 1120.0065 86.82929
## 4 1728.3812 63.90538
## 5 156.3692 413.87971
## 6 123.4769 443.71982转换为长矩阵
top6_t_with_group_melt <- gather(top6_t_with_group, key = "Gene", value = "Expr", -conditions, -Sample, -individual)
top6_t_with_group_melt## Sample conditions individual Gene Expr
## 1 trt_N052611 trt N052611 KCTD12 633.44616
## 2 trt_N061011 trt N061011 KCTD12 641.49582
## 3 trt_N080611 trt N080611 KCTD12 979.77576
## 4 trt_N61311 trt N61311 KCTD12 936.69481
## 5 untrt_N052611 untrt N052611 KCTD12 3978.04011
## 6 untrt_N061011 untrt N061011 KCTD12 4792.34174
## 7 untrt_N080611 untrt N080611 KCTD12 4416.15169
## 8 untrt_N61311 untrt N61311 KCTD12 4700.79369
## 9 trt_N052611 trt N052611 MAOA 4429.72011
## 10 trt_N061011 trt N061011 MAOA 3778.17351
## 11 trt_N080611 trt N080611 MAOA 4629.66529
## 12 trt_N61311 trt N61311 MAOA 4628.00860
## 13 untrt_N052611 untrt N052611 MAOA 452.99337
## 14 untrt_N061011 untrt N061011 MAOA 258.73279
## 15 untrt_N080611 untrt N080611 MAOA 516.63033
## 16 untrt_N61311 untrt N61311 MAOA 438.54451
## 17 trt_N052611 trt N052611 ARHGEF2 1441.99162
## 18 trt_N061011 trt N061011 ARHGEF2 1501.51509
## 19 trt_N080611 trt N080611 ARHGEF2 1464.59769
## 20 trt_N61311 trt N61311 ARHGEF2 1395.39850
## 21 untrt_N052611 untrt N052611 ARHGEF2 3105.78299
## 22 untrt_N061011 untrt N061011 ARHGEF2 2909.99043
## 23 untrt_N080611 untrt N080611 ARHGEF2 3094.51304
## 24 untrt_N61311 untrt N61311 ARHGEF2 3025.62334
## 25 trt_N052611 trt N052611 SPARCL1 1750.98786
## 26 trt_N061011 trt N061011 SPARCL1 2194.58930
## 27 trt_N080611 trt N080611 SPARCL1 1374.90745
## 28 trt_N61311 trt N61311 SPARCL1 2220.50867
## 29 untrt_N052611 untrt N052611 SPARCL1 102.58269
## 30 untrt_N061011 untrt N061011 SPARCL1 82.59042
## 31 untrt_N080611 untrt N080611 SPARCL1 80.00997
## 32 untrt_N61311 untrt N61311 SPARCL1 58.15705
## 33 trt_N052611 trt N052611 PER1 1230.25755
## 34 trt_N061011 trt N061011 PER1 1333.91208
## 35 trt_N080611 trt N080611 PER1 1120.00650
## 36 trt_N61311 trt N61311 PER1 1728.38117
## 37 untrt_N052611 untrt N052611 PER1 156.36920
## 38 untrt_N061011 untrt N061011 PER1 123.47689
## 39 untrt_N080611 untrt N080611 PER1 194.97497
## 40 untrt_N61311 untrt N61311 PER1 170.61639
## 41 trt_N052611 trt N052611 SLC6A9 56.89620
## 42 trt_N061011 trt N061011 SLC6A9 95.33916
## 43 trt_N080611 trt N080611 SLC6A9 86.82929
## 44 trt_N61311 trt N61311 SLC6A9 63.90538
## 45 untrt_N052611 untrt N052611 SLC6A9 413.87971
## 46 untrt_N061011 untrt N061011 SLC6A9 443.71982
## 47 untrt_N080611 untrt N080611 SLC6A9 365.47650
## 48 untrt_N61311 untrt N61311 SLC6A9 360.66314library(ggplot2)
ggplot(top6_t_with_group_melt, aes(x = Gene, y = Expr)) + geom_boxplot(aes(color = conditions)) + theme_classic()
7.4.6 配色
序列型颜色板适用于从低到高排序明显的数据,浅色数字小,深色数字大。

离散型颜色板适合带“正、负”的,对极值和中间值比较注重的数据。

分类型颜色板比较适合区分分类型的数据。

7.4.7 箱线图加统计分析
my_comparisons <- list(c("trt", "untrt"))
ggboxplot(top6_t_with_group, x = "conditions", y = "PER1",
title = "PER1", ylab = "Expression",
add = "jitter", # Add jittered points
#add = "dotplot",
fill = "conditions", palette = "Paired") +
stat_compare_means(comparisons = my_comparisons)
标记点来源的样本
my_comparisons <- list(c("trt", "untrt"))
ggboxplot(top6_t_with_group, x = "conditions", y = "PER1",
title = "PER1", ylab = "Expression",
add = "jitter", # Add jittered points
add.params = list(size = 0.1, jitter = 0.2), # Point size and the amount of jittering
label = "Sample", # column containing point labels
label.select = list(top.up = 2, top.down = 2),# Select some labels to display
font.label = list(size = 9, face = "italic"), # label font
repel = TRUE, # Avoid label text overplotting
fill = "conditions", palette = "Paired") +
stat_compare_means(comparisons = my_comparisons)
修改统计检验方法
my_comparisons <- list(c("trt", "untrt"))
ggboxplot(top6_t_with_group_log, x = "conditions", y = "PER1",
title = "PER1", ylab = "Expression",
add = "jitter", # Add jittered points
add.params = list(size = 0.1, jitter = 0.2), # Point size and the amount of jittering
label = "Sample", # column containing point labels
label.select = list(top.up = 2, top.down = 2),# Select some labels to display
font.label = list(size = 9, face = "italic"), # label font
repel = TRUE, # Avoid label text overplotting
fill = "conditions", palette = "Paired") +
stat_compare_means(comparisons = my_comparisons, method = "t.test", paired = T)
小提琴图
ggviolin(top6_t_with_group, x = "conditions", y = c("KCTD12","MAOA"),
ylab = "Expression", merge="flip",
color = "conditions", palette = "jco",
add = "boxplot"
# add = "median_iqr"
)
点带图(适合数据比较多时)
ggstripchart(top6_t_with_group, x = "conditions", y = c("KCTD12","MAOA"),
ylab = "Expression", combine=T,
color = "conditions", palette = "jco",
size = 0.1, jitter = 0.2,
add.params = list(color = "gray"),
# add = "boxplot"
add = "median_iqr")
7.5 通路内基因的比较
pathway <- read.table("h.all.v6.2.symbols.gmt.forGO", sep = "\t", row.names = NULL, header = T)
head(pathway)## ont gene
## 1 HALLMARK_TNFA_SIGNALING_VIA_NFKB JUNB
## 2 HALLMARK_TNFA_SIGNALING_VIA_NFKB CXCL2
## 3 HALLMARK_TNFA_SIGNALING_VIA_NFKB ATF3
## 4 HALLMARK_TNFA_SIGNALING_VIA_NFKB NFKBIA
## 5 HALLMARK_TNFA_SIGNALING_VIA_NFKB TNFAIP3
## 6 HALLMARK_TNFA_SIGNALING_VIA_NFKB PTGS2通路提取
# HALLMARK_HYPOXIA, HALLMARK_DNA_REPAIR, HALLMARK_P53_PATHWAY
target_pathway <- pathway[pathway$ont %in% c("HALLMARK_HYPOXIA", "HALLMARK_DNA_REPAIR", "HALLMARK_P53_PATHWAY"), ]
target_pathway <- droplevels.data.frame(target_pathway)
summary(target_pathway)## ont gene
## Length:550 Length:550
## Class :character Class :character
## Mode :character Mode :character## ont gene
## 201 HALLMARK_HYPOXIA PGK1
## 202 HALLMARK_HYPOXIA PDK1
## 203 HALLMARK_HYPOXIA GBE1
## 204 HALLMARK_HYPOXIA PFKL
## 205 HALLMARK_HYPOXIA ALDOA
## 206 HALLMARK_HYPOXIA ENO2表达矩阵提取
expr_with_gene <- expr
expr_with_gene$gene <- rownames(expr_with_gene)
target_pathway_with_expr <- left_join(target_pathway, expr_with_gene)
summary(target_pathway_with_expr)## ont gene untrt_N61311 untrt_N052611
## Length:550 Length:550 Min. : 0.0 Min. : 0.0
## Class :character Class :character 1st Qu.: 254.2 1st Qu.: 240.8
## Mode :character Mode :character Median : 781.3 Median : 784.1
## Mean : 2528.6 Mean : 2895.1
## 3rd Qu.: 1852.4 3rd Qu.: 1727.2
## Max. :212953.1 Max. :360796.2
## NA's :36 NA's :36
## untrt_N080611 untrt_N061011 trt_N61311 trt_N052611
## Min. : 0.0 Min. : 0.0 Min. : 0.0 Min. : 0.0
## 1st Qu.: 235.0 1st Qu.: 237.9 1st Qu.: 248.2 1st Qu.: 211.0
## Median : 734.9 Median : 764.2 Median : 766.6 Median : 723.2
## Mean : 2549.2 Mean : 2864.9 Mean : 2531.8 Mean : 2783.3
## 3rd Qu.: 1932.4 3rd Qu.: 1870.0 3rd Qu.: 1872.4 3rd Qu.: 1832.2
## Max. :258977.3 Max. :408573.1 Max. :210002.2 Max. :316009.1
## NA's :36 NA's :36 NA's :36 NA's :36
## trt_N080611 trt_N061011
## Min. : 0.0 Min. : 0.0
## 1st Qu.: 250.6 1st Qu.: 227.9
## Median : 739.3 Median : 746.0
## Mean : 2840.3 Mean : 3043.6
## 3rd Qu.: 1825.8 3rd Qu.: 1925.1
## Max. :225547.4 Max. :393843.7
## NA's :36 NA's :36移除通路中未检测到表达的基因
## ont gene untrt_N61311 untrt_N052611
## Length:514 Length:514 Min. : 0.0 Min. : 0.0
## Class :character Class :character 1st Qu.: 254.2 1st Qu.: 240.8
## Mode :character Mode :character Median : 781.3 Median : 784.1
## Mean : 2528.6 Mean : 2895.1
## 3rd Qu.: 1852.4 3rd Qu.: 1727.2
## Max. :212953.1 Max. :360796.2
## untrt_N080611 untrt_N061011 trt_N61311 trt_N052611
## Min. : 0.0 Min. : 0.0 Min. : 0.0 Min. : 0.0
## 1st Qu.: 235.0 1st Qu.: 237.9 1st Qu.: 248.2 1st Qu.: 211.0
## Median : 734.9 Median : 764.2 Median : 766.6 Median : 723.2
## Mean : 2549.2 Mean : 2864.9 Mean : 2531.8 Mean : 2783.3
## 3rd Qu.: 1932.4 3rd Qu.: 1870.0 3rd Qu.: 1872.4 3rd Qu.: 1832.2
## Max. :258977.3 Max. :408573.1 Max. :210002.2 Max. :316009.1
## trt_N080611 trt_N061011
## Min. : 0.0 Min. : 0.0
## 1st Qu.: 250.6 1st Qu.: 227.9
## Median : 739.3 Median : 746.0
## Mean : 2840.3 Mean : 3043.6
## 3rd Qu.: 1825.8 3rd Qu.: 1925.1
## Max. :225547.4 Max. :393843.7## ont gene untrt_N61311 untrt_N052611 untrt_N080611 untrt_N061011
## 1 HALLMARK_HYPOXIA PGK1 7567.398 7893.2150 6254.5945 5529.122
## 2 HALLMARK_HYPOXIA PDK1 1009.850 1042.4868 735.9359 673.208
## 3 HALLMARK_HYPOXIA GBE1 3859.557 1494.4120 3803.5627 3295.191
## 4 HALLMARK_HYPOXIA PFKL 3581.499 3018.0675 2789.4430 3084.570
## 5 HALLMARK_HYPOXIA ALDOA 19139.085 19587.3216 18089.5116 15519.899
## 6 HALLMARK_HYPOXIA ENO2 1964.796 979.5255 1041.4660 1288.837
## trt_N61311 trt_N052611 trt_N080611 trt_N061011
## 1 7595.0408 6969.6128 15011.823 6076.4392
## 2 419.6273 365.0062 1056.622 383.6163
## 3 4769.5464 2359.7150 4759.809 4296.5471
## 4 2867.2464 2599.5095 4399.403 3090.6701
## 5 16388.1123 13949.5659 22630.701 14374.3437
## 6 1303.5671 766.9436 1473.336 892.4621转换宽矩阵为长矩阵
target_pathway_with_expr_long <- target_pathway_with_expr %>% gather(key = "Sample", value = "Expr", -ont, -gene)
head(target_pathway_with_expr_long)## ont gene Sample Expr
## 1 HALLMARK_HYPOXIA PGK1 untrt_N61311 7567.398
## 2 HALLMARK_HYPOXIA PDK1 untrt_N61311 1009.850
## 3 HALLMARK_HYPOXIA GBE1 untrt_N61311 3859.557
## 4 HALLMARK_HYPOXIA PFKL untrt_N61311 3581.499
## 5 HALLMARK_HYPOXIA ALDOA untrt_N61311 19139.085
## 6 HALLMARK_HYPOXIA ENO2 untrt_N61311 1964.796合并样本信息
metadata$Sample <- rownames(metadata)
target_pathway_with_expr_conditions_long <- target_pathway_with_expr_long %>% left_join(metadata, by = "Sample")
head(target_pathway_with_expr_conditions_long)## ont gene Sample Expr conditions individual
## 1 HALLMARK_HYPOXIA PGK1 untrt_N61311 7567.398 untrt N61311
## 2 HALLMARK_HYPOXIA PDK1 untrt_N61311 1009.850 untrt N61311
## 3 HALLMARK_HYPOXIA GBE1 untrt_N61311 3859.557 untrt N61311
## 4 HALLMARK_HYPOXIA PFKL untrt_N61311 3581.499 untrt N61311
## 5 HALLMARK_HYPOXIA ALDOA untrt_N61311 19139.085 untrt N61311
## 6 HALLMARK_HYPOXIA ENO2 untrt_N61311 1964.796 untrt N61311再次画点带图 (也不太好看)
ggstripchart(target_pathway_with_expr_conditions_long, x = "conditions", y = "Expr",
ylab = "Expression", combine=F,
color = "conditions", palette = "jco",
size = 0.1, jitter = 0.2,
facet.by = "ont",
add.params = list(color = "gray"),
# add = "boxplot"
add = "median_iqr")
表达数据log转换(减小高表达基因的影响)
target_pathway_with_expr_conditions_long$logExpr <- log2(target_pathway_with_expr_conditions_long$Expr + 1)
ggstripchart(target_pathway_with_expr_conditions_long, x = "conditions", y = "logExpr",
ylab = "Expression", combine=F,
color = "conditions", palette = "jco",
size = 0.1, jitter = 0.2,
facet.by = "ont",
add.params = list(color = "gray"),
# add = "boxplot"
add = "median_iqr")
## ont gene Sample Expr conditions individual logExpr
## 1 HALLMARK_HYPOXIA PGK1 untrt_N61311 7567.398 untrt N61311 12.885772
## 2 HALLMARK_HYPOXIA PDK1 untrt_N61311 1009.850 untrt N61311 9.981353
## 3 HALLMARK_HYPOXIA GBE1 untrt_N61311 3859.557 untrt N61311 11.914593
## 4 HALLMARK_HYPOXIA PFKL untrt_N61311 3581.499 untrt N61311 11.806750
## 5 HALLMARK_HYPOXIA ALDOA untrt_N61311 19139.085 untrt N61311 14.224310
## 6 HALLMARK_HYPOXIA ENO2 untrt_N61311 1964.796 untrt N61311 10.940898提取P53通路进行后续分析
HALLMARK_P53_PATHWAY = target_pathway_with_expr_conditions_long[target_pathway_with_expr_conditions_long$ont=="HALLMARK_P53_PATHWAY",]
ggstripchart(HALLMARK_P53_PATHWAY, x = "conditions", y = "logExpr",
title = "HALLMARK_P53_PATHWAY",
ylab = "Expression",
color = "conditions", palette = "jco",
size = 0.1, jitter = 0.2,
add.params = list(color = "gray"),
# add = "boxplot"
add = "median_iqr")
ggdotplot(HALLMARK_P53_PATHWAY, x = "conditions", y = "logExpr",
title = "HALLMARK_P53_PATHWAY",
ylab = "Expression",
color = "conditions", palette = "jco",
fill = "white",
binwidth = 0.1,
add.params = list(size = 0.9),
# add = "boxplot"
add = "median_iqr")
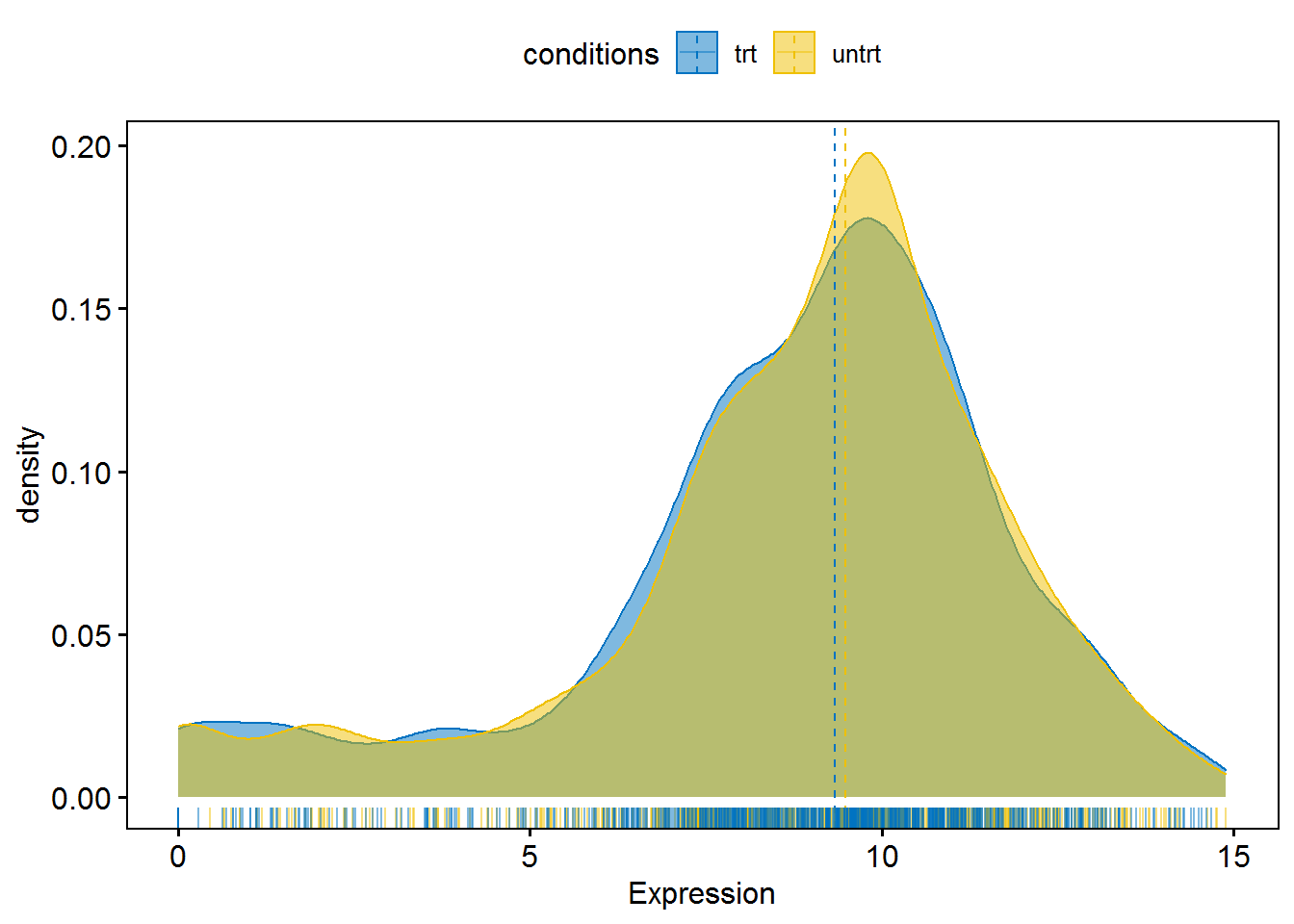
7.5.1 密度图
ggdensity(HALLMARK_P53_PATHWAY,
x="logExpr",
y = "..density..",
combine = TRUE, # Combine the 3 plots
xlab = "Expression",
add = "median", # Add median line.
rug = TRUE, # Add marginal rug
color = "conditions",
fill = "conditions",
palette = "jco"
)
## Sample conditions individual KCTD12 MAOA ARHGEF2 SPARCL1
## 1 trt_N052611 trt N052611 633.4462 4429.7201 1441.992 1750.98786
## 2 trt_N061011 trt N061011 641.4958 3778.1735 1501.515 2194.58930
## 3 trt_N080611 trt N080611 979.7758 4629.6653 1464.598 1374.90745
## 4 trt_N61311 trt N61311 936.6948 4628.0086 1395.398 2220.50867
## 5 untrt_N052611 untrt N052611 3978.0401 452.9934 3105.783 102.58269
## 6 untrt_N061011 untrt N061011 4792.3417 258.7328 2909.990 82.59042
## PER1 SLC6A9
## 1 1230.2575 56.89620
## 2 1333.9121 95.33916
## 3 1120.0065 86.82929
## 4 1728.3812 63.90538
## 5 156.3692 413.87971
## 6 123.4769 443.71982top6_t_with_group_long = top6_t_with_group %>% gather(key = "Gene", value = "Expr", -conditions, -Sample, -individual)
top6_t_with_group_long$conditions <- as.factor(top6_t_with_group_long$conditions)
head(top6_t_with_group_long)## Sample conditions individual Gene Expr
## 1 trt_N052611 trt N052611 KCTD12 633.4462
## 2 trt_N061011 trt N061011 KCTD12 641.4958
## 3 trt_N080611 trt N080611 KCTD12 979.7758
## 4 trt_N61311 trt N61311 KCTD12 936.6948
## 5 untrt_N052611 untrt N052611 KCTD12 3978.0401
## 6 untrt_N061011 untrt N061011 KCTD12 4792.34177.6 ggstatsplot绘图和统计分析
箱线图
library(ggstatsplot)
ggstatsplot::ggwithinstats(
data = top6_t_with_group,
x = conditions,
y = PER1,
sort = "descending", # ordering groups along the x-axis based on
sort.fun = median, # values of `y` variable
pairwise.comparisons = TRUE,
pairwise.display = "s",
pairwise.annotation = "p",
title = "PER1",
caption = "PER1 compare",
ggstatsplot.layer = FALSE,
messages = FALSE
)
## ont gene Sample Expr conditions individual logExpr
## 1 HALLMARK_HYPOXIA PGK1 untrt_N61311 7567.398 untrt N61311 12.885772
## 2 HALLMARK_HYPOXIA PDK1 untrt_N61311 1009.850 untrt N61311 9.981353
## 3 HALLMARK_HYPOXIA GBE1 untrt_N61311 3859.557 untrt N61311 11.914593
## 4 HALLMARK_HYPOXIA PFKL untrt_N61311 3581.499 untrt N61311 11.806750
## 5 HALLMARK_HYPOXIA ALDOA untrt_N61311 19139.085 untrt N61311 14.224310
## 6 HALLMARK_HYPOXIA ENO2 untrt_N61311 1964.796 untrt N61311 10.940898## ont gene Sample Expr conditions individual
## 322 HALLMARK_P53_PATHWAY CDKN1A untrt_N61311 14406.1316 untrt N61311
## 323 HALLMARK_P53_PATHWAY BTG2 untrt_N61311 1163.7198 untrt N61311
## 324 HALLMARK_P53_PATHWAY MDM2 untrt_N61311 3614.5324 untrt N61311
## 325 HALLMARK_P53_PATHWAY CCNG1 untrt_N61311 5749.1367 untrt N61311
## 326 HALLMARK_P53_PATHWAY FAS untrt_N61311 1029.4007 untrt N61311
## 327 HALLMARK_P53_PATHWAY TOB1 untrt_N61311 829.7721 untrt N61311
## logExpr
## 322 13.814496
## 323 10.185767
## 324 11.819992
## 325 12.489381
## 326 10.008990
## 327 9.698309library(ggstatsplot)
ggstatsplot::ggwithinstats(
data = HALLMARK_P53_PATHWAY,
x = conditions,
y = logExpr,
sort = "descending", # ordering groups along the x-axis based on
sort.fun = median, # values of `y` variable
pairwise.comparisons = TRUE,
pairwise.display = "s",
pairwise.annotation = "p",
title = "HALLMARK_P53_PATHWAY",
path.point = F,
ggtheme = ggthemes::theme_fivethirtyeight(),
ggstatsplot.layer = FALSE,
messages = FALSE
)
library(ggstatsplot)
ggstatsplot::grouped_ggwithinstats(
data = target_pathway_with_expr_conditions_long,
x = conditions,
y = logExpr,
grouping.var = ont,
xlab = "Condition",
ylab = "CEMIP expression",
path.point = F,
palette = "Set1", # R color brewer
ggstatsplot.layer = FALSE,
messages = FALSE
)
ggstatsplot::grouped_ggwithinstats(data = top6_t_with_group_long, x = conditions, y = Expr, xlab = "Condition", ylab = "CEMIP expression",
grouping.var = Gene, ggstatsplot.layer = FALSE, messages = FALSE)
## untrt_N61311 untrt_N052611 untrt_N080611 untrt_N061011 trt_N61311
## FN1 245667.66 427435.1 221687.51 371144.2 240187.24
## DCN 212953.14 360796.2 258977.30 408573.1 210002.18
## CEMIP 40996.34 137783.1 53813.92 91066.8 62301.12
## CCDC80 137229.15 232772.2 86258.13 212237.3 136730.76
## IGFBP5 77812.65 288609.2 210628.87 168067.4 96021.74
## COL1A1 146450.41 127367.3 152281.50 140861.1 62358.64
## trt_N052611 trt_N080611 trt_N061011
## FN1 450103.21 280226.19 376518.23
## DCN 316009.14 225547.39 393843.74
## CEMIP 223111.85 212724.84 157919.47
## CCDC80 226070.89 124634.56 236237.81
## IGFBP5 217439.21 162677.38 168387.36
## COL1A1 53800.47 69160.97 51044.067.6.1 散点图
ggstatsplot::ggscatterstats(data = expr, x = untrt_N61311, y = untrt_N052611, xlab = "untrt_N61311", ylab = "untrt_N052611",
title = "Sample correlation", messages = FALSE)
ggstatsplot::ggscatterstats(
data = log2(expr+1),
x = untrt_N61311,
y = trt_N61311,
xlab = "untrt_N61311",
ylab = "trt_N61311",
title = "Sample correlation",
#marginal.type = "density", # type of marginal distribution to be displayed
messages = FALSE
)
7.6.2 相关性图
7.6.2.1 基因共表达
## KCTD12 MAOA ARHGEF2 SPARCL1 PER1 SLC6A9
## KCTD12 1.0000000 -0.9792624 0.9799663 -0.9619660 -0.9529732 0.9772852
## MAOA -0.9792624 1.0000000 -0.9897706 0.9406196 0.9614877 -0.9871408
## ARHGEF2 0.9799663 -0.9897706 1.0000000 -0.9628750 -0.9660416 0.9791535
## SPARCL1 -0.9619660 0.9406196 -0.9628750 1.0000000 0.9853858 -0.9510121
## PER1 -0.9529732 0.9614877 -0.9660416 0.9853858 1.0000000 -0.9615253
## SLC6A9 0.9772852 -0.9871408 0.9791535 -0.9510121 -0.9615253 1.0000000
## KCTD12 MAOA ARHGEF2 SPARCL1 PER1 SLC6A9
## KCTD12 1.00 -0.98 0.98 -0.96 -0.95 0.98
## MAOA -0.98 1.00 -0.99 0.94 0.96 -0.99
## ARHGEF2 0.98 -0.99 1.00 -0.96 -0.97 0.98
## SPARCL1 -0.96 0.94 -0.96 1.00 0.99 -0.95
## PER1 -0.95 0.96 -0.97 0.99 1.00 -0.96
## SLC6A9 0.98 -0.99 0.98 -0.95 -0.96 1.00
##
## n= 8
##
##
## P
## KCTD12 MAOA ARHGEF2 SPARCL1 PER1 SLC6A9
## KCTD12 0e+00 0e+00 1e-04 3e-04 0e+00
## MAOA 0e+00 0e+00 5e-04 1e-04 0e+00
## ARHGEF2 0e+00 0e+00 1e-04 0e+00 0e+00
## SPARCL1 1e-04 5e-04 1e-04 0e+00 3e-04
## PER1 3e-04 1e-04 0e+00 0e+00 1e-04
## SLC6A9 0e+00 0e+00 0e+00 3e-04 1e-04## KCTD12 MAOA ARHGEF2 SPARCL1 PER1 SLC6A9
## untrt_N61311 4700.7937 438.5445 3025.623 58.15705 170.6164 360.66314
## untrt_N052611 3978.0401 452.9934 3105.783 102.58269 156.3692 413.87971
## untrt_N080611 4416.1517 516.6303 3094.513 80.00997 194.9750 365.47650
## untrt_N061011 4792.3417 258.7328 2909.990 82.59042 123.4769 443.71982
## trt_N61311 936.6948 4628.0086 1395.398 2220.50867 1728.3812 63.90538
## trt_N052611 633.4462 4429.7201 1441.992 1750.98786 1230.2575 56.89620ggstatsplot::ggcorrmat(
data = top6_t,
corr.method = "robust", # correlation method
sig.level = 0.0001, # threshold of significance
p.adjust.method = "holm", # p-value adjustment method for multiple comparisons
# cor.vars = c(sleep_rem, awake:bodywt), # a range of variables can be selected
# cor.vars.names = c(
# "REM sleep", # variable names
# "time awake",
# "brain weight",
# "body weight"
# ),
matrix.type = "upper", # type of visualization matrix
palette = "Set2",
#colors = c("#B2182B", "white", "#4D4D4D"),
title = "Correlalogram for mammals sleep dataset",
subtitle = "sleep units: hours; weight units: kilograms"
)
7.6.2.2 样品相关性
top100 <- head(expr,100)
ggstatsplot::ggcorrmat(
data = top100,
corr.method = "robust", # correlation method
sig.level = 0.05, # threshold of significance
p.adjust.method = "holm", # p-value adjustment method for multiple comparisons
# cor.vars = c(sleep_rem, awake:bodywt), # a range of variables can be selected
# cor.vars.names = c(
# "REM sleep", # variable names
# "time awake",
# "brain weight",
# "body weight"
# ),
matrix.type = "upper", # type of visualization matrix
palette = "Set2"
#colors = c("#B2182B", "white", "#4D4D4D"),
)