4 基于RandomForest学习Caret标准化流程
Caret 是一个试图标准化机器学习过程的一个包。Caret 对 R 中最常用的机器学习方法 (目前支持238个R包,具体列表见https://topepo.github.io/caret/available-models.html,也可以自己根据已有方法的模板快速添加新的方法)提供了统一的接口。
- 进行数据预处理
- 实现机器学习方法流程化模型构建
- 通过参数组合和交叉验证评估模型的参数
- 选择最优模型
- 评估模型性能
4.1 基于Caret和RandomForest包进行随机森林分析的一般步骤
4.1.1 Caret构建机器学习流程的一般步骤
Caret依赖trainControl函数设置交叉验证参数,train函数具体训练和评估模型。首先是选择一系列需要评估的参数和参数值的组合,然后设置重采样评估方式,循环训练模型评估结果、计算模型的平均性能,根据设定的度量值选择最好的模型参数组合,使用全部训练集和最优参数组合完成模型的最终训练。

Figure 4.1: Caret的工作流程. https://topepo.github.io/caret/model-training-and-tuning.html
4.1.2 基于Caret和RandomForest包进行随机森林分析的一般步骤
createDataPartition是拆分数据为训练集和测试集的函数。对于分类数据,按照每个类的大小成比例拆分;如果是回归数据,则先把响应值分为n个区间,再成比例拆分。
library(caret)
# 拆分数据为测试集和训练集
seed <- 1
set.seed(seed)
train_index <- createDataPartition(metadata[[group]], p=0.75, list=F)
train_data <- expr_mat[train_index,]
train_data_group <- metadata[[group]][train_index]
test_data <- expr_mat[-train_index,]
test_data_group <- metadata[[group]][-train_index]
# Create model with default parameters
# repeats可增加
seedL = generateTrainControlSeeds(number_k_fold=10, repeat_k_fold=3, num_parameters=100)
trControl <- trainControl(method="repeatedcv", number=10, repeats=3, seeds=seedL)
# train model
if(file.exists('rda/rf_default.rda')){
rf_default <- readRDS("rda/rf_default.rda")
} else {
# 设置随机数种子,使得结果可重复
seed <- 1
set.seed(seed)
# method = rf: 表示使用randomForest包的随机森林算法
# 各个包对应的method可通过链接获得
# https://topepo.github.io/caret/train-models-by-tag.html
rf_default <- train(x=train_data, y=train_data_group, method="rf",
trControl=trControl)
saveRDS(rf_default, "rda/rf_default.rda")
}
print(rf_default)## Random Forest
##
## 59 samples
## 7070 predictors
## 2 classes: 'DLBCL', 'FL'
##
## No pre-processing
## Resampling: Cross-Validated (10 fold, repeated 3 times)
## Summary of sample sizes: 53, 53, 54, 53, 53, 54, ...
## Resampling results across tuning parameters:
##
## mtry Accuracy Kappa
## 2 0.7612698 0.05238095
## 118 0.8680952 0.55866947
## 7069 0.9065079 0.68897759
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was mtry = 7069.精确性随默认调参的变化
plot(rf_default)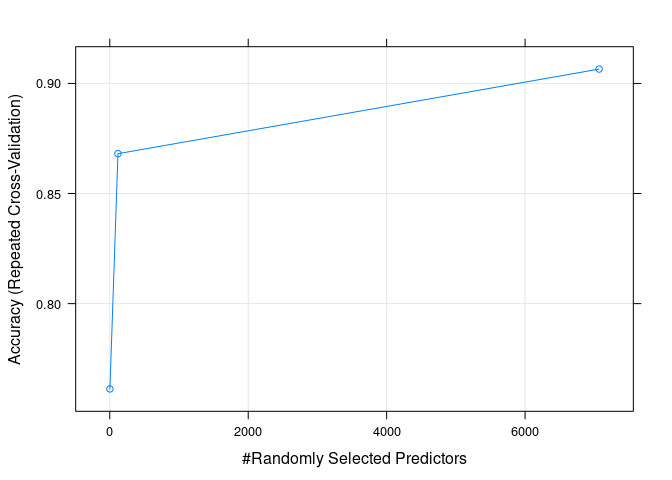
Figure 4.2: 精确性随默认调参的变化
Kappa值随默认调参的变化
# ?plot.train查看更多使用信息
plot(rf_default, metric="Kappa")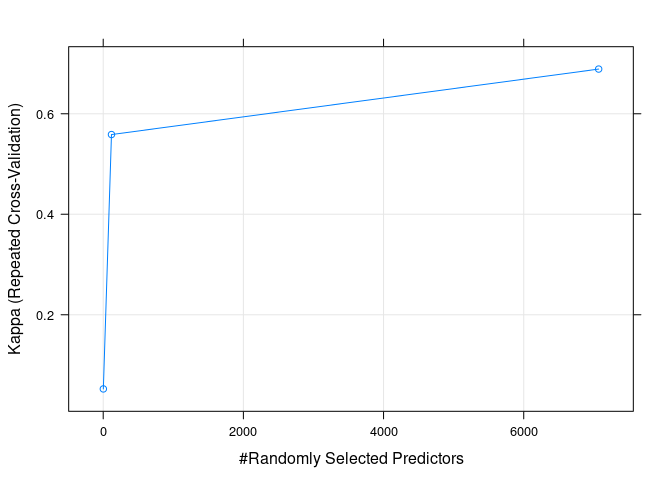
Figure 4.3: Kappa值随默认调参的变化
#str(rf_default)4.1.3 Caret比较不同算法的性能
Caret包的流程统一性在这就体现出来了,我之前没有看过ranger和parRF包,也不知道他们的具体使用。但我可以借助Caret很快地用他们构建一个初步模型,并与randomForest的结果进行比较。
# Too slow
# RRF: Regularized Random Forest
# 约2个小时,普通笔记本
#确认Cross-validation分割数据的种子一致
trControl$seeds <- rf_default$control$seeds
if(file.exists('rda/RRF_default.rda')){
RRF_default <- readRDS("rda/RRF_default.rda")
} else {
seed = 1
set.seed(seed)
RRF_default <- train(x=train_data, y=train_data_group, method="RRF",
trControl=trControl)
saveRDS(RRF_default, "rda/RRF_default.rda")
}
RRF_default## Regularized Random Forest
##
## 59 samples
## 7070 predictors
## 2 classes: 'DLBCL', 'FL'
##
## No pre-processing
## Resampling: Cross-Validated (10 fold, repeated 3 times)
## Summary of sample sizes: 53, 53, 54, 53, 53, 54, ...
## Resampling results across tuning parameters:
##
## mtry coefReg coefImp Accuracy Kappa
## 2 0.010 0.0 0.6725397 0.08144749
## 2 0.010 0.5 0.6752381 0.17248645
## 2 0.010 1.0 0.7866667 0.36026123
## 2 0.505 0.0 0.7034921 0.10858798
## 2 0.505 0.5 0.6841270 0.11599992
## 2 0.505 1.0 0.7230159 0.19266658
## 2 1.000 0.0 0.8514286 0.53971756
## 2 1.000 0.5 0.7974603 0.38003715
## 2 1.000 1.0 0.7369841 0.23509337
## 118 0.010 0.0 0.7352381 0.26154613
## 118 0.010 0.5 0.7666667 0.34993464
## 118 0.010 1.0 0.8288889 0.50908943
## 118 0.505 0.0 0.6885714 0.15668942
## 118 0.505 0.5 0.7855556 0.35756504
## 118 0.505 1.0 0.8538095 0.60203996
## 118 1.000 0.0 0.9069841 0.69492997
## 118 1.000 0.5 0.8847619 0.66954228
## 118 1.000 1.0 0.8017460 0.44832381
## 7069 0.010 0.0 0.7700000 0.34325805
## 7069 0.010 0.5 0.7073016 0.19615064
## 7069 0.010 1.0 0.7746032 0.36585509
## 7069 0.505 0.0 0.7038095 0.27730116
## 7069 0.505 0.5 0.7438095 0.29364891
## 7069 0.505 1.0 0.7557143 0.32099191
## 7069 1.000 0.0 0.8950794 0.65334734
## 7069 1.000 0.5 0.8188889 0.49086796
## 7069 1.000 1.0 0.7557143 0.35696593
##
## Accuracy was used to select the optimal model using the largest value.
## The final values used for the model were mtry = 118, coefReg = 1 and coefImp
## = 0.ranger是random forest的快速版本,特别适用于高维数据,支持分类、回归和生存分析。
# ranger
# ranger is a fast implementation of random forests (Breiman 2001) or recursive partitioning, particularly suited for high dimensional data. Classification, regression, and survival forests are supported. Classification and regression forests are implemented as in the original Random Forest (Breiman 2001), survival forests as in Random Survival Forests (Ishwaran et al. 2008).
# 10 分钟
trControl$seeds <- rf_default$control$seeds
if(file.exists('rda/ranger_default.rda')){
ranger_default <- readRDS("rda/ranger_default.rda")
} else {
set.seed(1)
ranger_default <- train(x=train_data, y=train_data_group, method="ranger",
trControl=trControl)
saveRDS(ranger_default, "rda/ranger_default.rda")
}
ranger_default## Random Forest
##
## 59 samples
## 7070 predictors
## 2 classes: 'DLBCL', 'FL'
##
## No pre-processing
## Resampling: Cross-Validated (10 fold, repeated 3 times)
## Summary of sample sizes: 53, 53, 54, 53, 53, 54, ...
## Resampling results across tuning parameters:
##
## mtry splitrule Accuracy Kappa
## 2 gini 0.7546032 0.01904762
## 2 extratrees 0.7490476 0.00000000
## 118 gini 0.8733333 0.56096639
## 118 extratrees 0.8041270 0.21540616
## 7069 gini 0.8950794 0.65334734
## 7069 extratrees 0.9128571 0.72001401
##
## Tuning parameter 'min.node.size' was held constant at a value of 1
## Accuracy was used to select the optimal model using the largest value.
## The final values used for the model were mtry = 7069, splitrule =
## extratrees and min.node.size = 1.parRF是并行随机森林算法。
# parRF: Parallel Random Forest
# 0.67s
trControl$seeds <- rf_default$control$seeds
if(file.exists('rda/parRF_default.rda')){
parRF_default <- readRDS("rda/parRF_default.rda")
} else {
# 其它部分也都可以使用这段并行代码加速
library(doParallel)
cl <- makePSOCKcluster(2)
registerDoParallel(cl)
# 并行代码初始化结束
set.seed(1)
parRF_default <- train(x=train_data, y=train_data_group, method="parRF",
trControl=trControl)
## 并行cluster关闭
stopCluster(cl)
saveRDS(parRF_default, "rda/parRF_default.rda")
}
parRF_default## Parallel Random Forest
##
## 59 samples
## 7070 predictors
## 2 classes: 'DLBCL', 'FL'
##
## No pre-processing
## Resampling: Cross-Validated (10 fold, repeated 3 times)
## Summary of sample sizes: 53, 53, 54, 53, 53, 54, ...
## Resampling results across tuning parameters:
##
## mtry Accuracy Kappa
## 2 0.7546032 0.01904762
## 118 0.8622222 0.51334734
## 7069 0.8884127 0.62413165
##
## Accuracy was used to select the optimal model using the largest value.
## The final value used for the model was mtry = 7069.合并四个包的模型结果
resamps <- resamples(list(RRF = RRF_default,
rf = rf_default,
parRF = parRF_default,
ranger = ranger_default))
resamps##
## Call:
## resamples.default(x = list(RRF = RRF_default, rf = rf_default, parRF
## = parRF_default, ranger = ranger_default))
##
## Models: RRF, rf, parRF, ranger
## Number of resamples: 30
## Performance metrics: Accuracy, Kappa
## Time estimates for: everything, final model fit比较绘制四个包的性能差异
theme1 <- trellis.par.get()
theme1$plot.symbol$col = rgb(.2, .2, .2, .4)
theme1$plot.symbol$pch = 16
theme1$plot.line$col = rgb(1, 0, 0, .7)
theme1$plot.line$lwd <- 2
trellis.par.set(theme1)
# 这个图看着很奇怪,Kappa值高估
bwplot(resamps)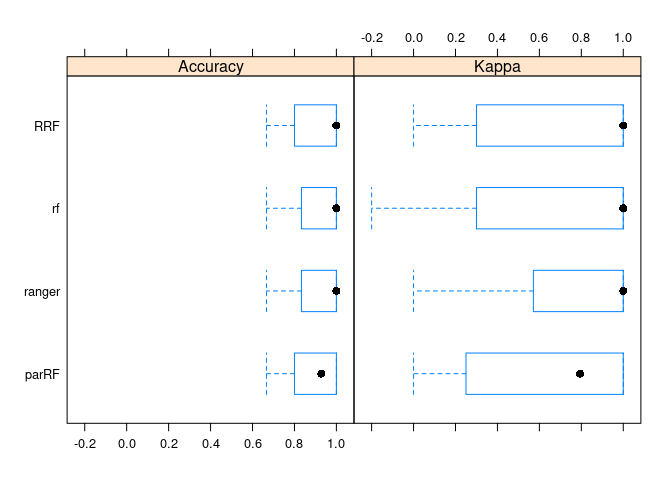
Figure 4.4: 三个包性能相当
summary(resamps)##
## Call:
## summary.resamples(object = resamps)
##
## Models: RRF, rf, parRF, ranger
## Number of resamples: 30
##
## Accuracy
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## RRF 0.6666667 0.8083333 1.0000000 0.9069841 1 1 0
## rf 0.6666667 0.8333333 1.0000000 0.9065079 1 1 0
## parRF 0.6666667 0.8000000 0.9285714 0.8884127 1 1 0
## ranger 0.6666667 0.8333333 1.0000000 0.9128571 1 1 0
##
## Kappa
## Min. 1st Qu. Median Mean 3rd Qu. Max. NA's
## RRF 0.0 0.3000000 1.0000000 0.6949300 1 1 0
## rf -0.2 0.3678571 1.0000000 0.6889776 1 1 0
## parRF 0.0 0.2625000 0.7941176 0.6241317 1 1 0
## ranger 0.0 0.5714286 1.0000000 0.7200140 1 1 0dotplot(resamps)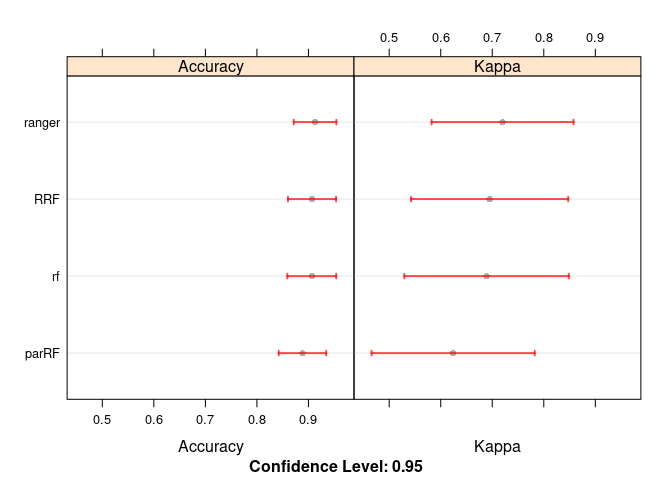
统计检验评估模型之间差异是否显著
diff.value <- diff(resamps)
summary(diff.value)##
## Call:
## summary.diff.resamples(object = diff.value)
##
## p-value adjustment: bonferroni
## Upper diagonal: estimates of the difference
## Lower diagonal: p-value for H0: difference = 0
##
## Accuracy
## RRF rf parRF ranger
## RRF 0.0004762 0.0185714 -0.0058730
## rf 1.0000 0.0180952 -0.0063492
## parRF 1.0000 1.0000 -0.0244444
## ranger 1.0000 1.0000 0.4828
##
## Kappa
## RRF rf parRF ranger
## RRF 0.005952 0.070798 -0.025084
## rf 1.0000 0.064846 -0.031036
## parRF 1.0000 1.0000 -0.095882
## ranger 1.0000 1.0000 0.50014.1.4 Caret训练最终模型
if(file.exists('rda/rf_finaltest.rda')){
rf_final <- readRDS("rda/rf_finaltest.rda")
} else {
# method="none" 不再抽样,用全部数据训练模型
trControl <- trainControl(method="none", classProbs = T)
# 设置随机数种子,使得结果可重复
seed <- 1
set.seed(seed)
rf_final <- train(x=train_data, y=train_data_group, method="rf",
# 用最好的模型的调参值
tuneGrid = rf_default$bestTune,
trControl=trControl)
saveRDS(rf_final, "rda/rf_finaltest.rda")
}
print(rf_final)## Random Forest
##
## 59 samples
## 7070 predictors
## 2 classes: 'DLBCL', 'FL'
##
## No pre-processing
## Resampling: None4.1.5 基于模型对测试集进行预测
# ?predict.train
# type: raw (返回预测的类或回归的数值) or prob (返回分类到每个类的概率)
predict(rf_final, newdata=head(test_data))## [1] DLBCL DLBCL DLBCL DLBCL DLBCL DLBCL
## Levels: DLBCL FL虽然都能被分类到DLBCL,但分类概率是不同的,如DLBCL_16就在边缘徘徊。
# ?predict.train
# type: raw (返回预测的类或回归的数值) or prob (返回分类到每个类的概率)
predict(rf_final, newdata=head(test_data), type="prob")## DLBCL FL
## DLBCL_2 0.960 0.040
## DLBCL_5 0.934 0.066
## DLBCL_11 0.742 0.258
## DLBCL_13 0.892 0.108
## DLBCL_16 0.866 0.134
## DLBCL_17 0.838 0.162获得模型结果评估矩阵(confusion matrix)
predictions <- predict(rf_final, newdata=test_data)
confusionMatrix(predictions, test_data_group)## Confusion Matrix and Statistics
##
## Reference
## Prediction DLBCL FL
## DLBCL 14 2
## FL 0 2
##
## Accuracy : 0.8889
## 95% CI : (0.6529, 0.9862)
## No Information Rate : 0.7778
## P-Value [Acc > NIR] : 0.2022
##
## Kappa : 0.6087
##
## Mcnemar's Test P-Value : 0.4795
##
## Sensitivity : 1.0000
## Specificity : 0.5000
## Pos Pred Value : 0.8750
## Neg Pred Value : 1.0000
## Prevalence : 0.7778
## Detection Rate : 0.7778
## Detection Prevalence : 0.8889
## Balanced Accuracy : 0.7500
##
## 'Positive' Class : DLBCL
## 绘制ROC曲线
prediction_prob <- predict(rf_final, newdata=test_data, type="prob")
library(pROC)
roc <- roc(test_data_group, prediction_prob[,1])
#roc <- roc(test_data_group, factor(predictions, ordered=T))
roc##
## Call:
## roc.default(response = test_data_group, predictor = prediction_prob[, 1])
##
## Data: prediction_prob[, 1] in 14 controls (test_data_group DLBCL) > 4 cases (test_data_group FL).
## Area under the curve: 0.9821ROC_data <- data.frame(FPR = 1- roc$specificities, TPR=roc$sensitivities)
ROC_data <- ROC_data[with(ROC_data, order(FPR,TPR)),]
p <- ggplot(data=ROC_data, mapping=aes(x=FPR, y=TPR)) +
geom_step(color="red", size=1, direction = "vh") +
geom_segment(aes(x=0, xend=1, y=0, yend=1)) + theme_classic() +
xlab("False positive rate") +
ylab("True positive rate") + coord_fixed(1) + xlim(0,1) + ylim(0,1) +
annotate('text', x=0.5, y=0.25, label=paste('AUC=', round(roc$auc,2)))
p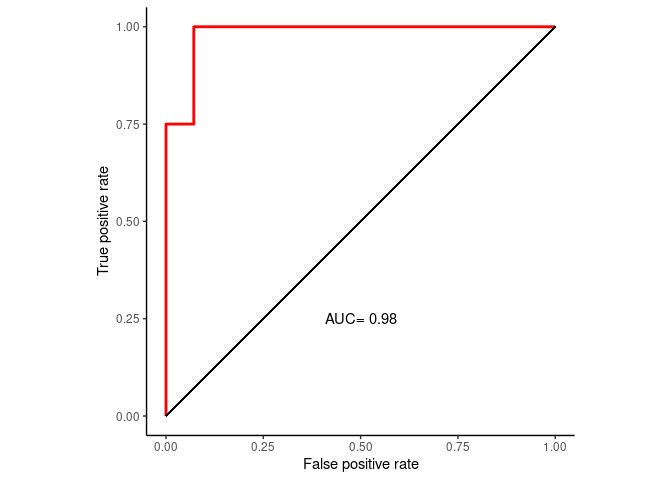
4.2 Caret模型训练和调参更多参数解读
4.2.1 trainControl函数控制参数
trainControl函数用于定义train函数运行的一些参数,如交叉验证方式、模型评估函数、模型选择标准、调参方式等。
部分参数解释如下:
- method: 重采样方法”boot”, “cv”, “LOOCV”, “LGOCV”, “repeatedcv”, “timeslice”, “none” and “oob”。
oob只适用于random forest, bagged trees, bagged earth, bagged flexible discriminant analysis, 或 conditional tree forest models. ’none’表示不进行重采样。repeatedcv是K-fold交叉验证重复多次。 - number: K-fold中的
K或boot中的迭代次数。 - repeats: 只适用于
repeatedcv,重复K-fold`交叉验证数次 method = "repeatedcv",number = 10和repeats = 3表示完整重复3次10-fold交叉验证。- search:选项是
grid(矩阵调参)或random(随机调参) - summaryFunction: 计算模型性能矩阵的函数,默认是
defaultSummary;twoClassSummary用来计算敏感性、特异性和AUC。 - sampling: 在数据不平衡时的抽平方式。
down-sampling把所有类的样品抽取到与最小的类样品数一致;up-sampling有放回的抽取使得所有类的样品与最大的类样品数一致。实际上是否随机采样和用什么采样方法获得的结果一般是一致的。https://topepo.github.io/caret/subsampling-for-class-imbalances.html - classProbs: 是(TRUE)否(FALSE)需要计算验证集中样品的分类概率。
- seed: 通常在运行
train程序前运行set.seed(1)就可以保证结果的可重复性。但在使用并行计算时,根据技术不同,可能需要额外设置此参数。 - index: 自定义训练集
# Do not run, only for an example
control <- trainControl(method="repeatedcv", number=10, repeats=3,
search="grid", selectionFunction ="tolerance(tol=2)")4.2.2 train函数控制参数
metric: 模型度量标准;回归分析默认使用RMSE,R-square和MAE (mean absolute error), 分类问题默认使用Accuracy和Kappa;如果指定ROC值,需要指定trainControl的summaryFunction=twoClassSummary(函数名无需引号),并且classProbs = TRUE。selectionFunction: Caret自带了3个函数,best: 按照设定的metric值选择评分最高或偏差最小的模型;oneSE选择最简单的其性能与最好模型的性能相差在一个标准差之内的模型(同时照顾性能和避免过拟合);tolerance选择与最好的模型相差在给定阈值范围内的最简单模型。阈值为容忍度,计算方式为 $ (x-x_{best})/x_{best} * 100$。selectionFunction ="tolerance(tol=2)"表示允许2%的性能降低。tuneGrid: 接受一个包含参数组合的数据框。列的名字是所用的机器学习方法函数可接受的参数名字。getModelInfo()$rf$parameters可获得方法rf的所有可调参数。
# Do not run, only for an example
# # 模型评估标准
metric <- "Accuracy"
control <- trainControl(method="repeatedcv", number=10, repeats=3,
search="grid", selectionFunction ="tolerance(tol=2)")
set.seed(seed)
tunegrid <- expand.grid(.mtry=c(1:15))
# 设置构建决策树每步决策用到的变量数
# 这里是默认参数
mtry <- sqrt(ncol(expr_mat))
# 构建调参矩阵
# tuneGrid可以接受一个包含参数组合的数据框。列的名字是所用的机器学习方法函数可接受的参数名字.
# getModelInfo()$rf$parameters 可获得方法rf的所有可调参数
tunegrid <- expand.grid(mtry=mtry)
rf_gridsearch <- train(Class~., data=dataset, method="rf", metric=metric,
tuneGrid=tunegrid, trControl=control, selectionFunction)
print(rf_gridsearch)
plot(rf_gridsearch)4.3 References
- 官方文档 https://topepo.github.io/caret/model-training-and-tuning.html
- 部分中文解释 https://blog.csdn.net/weixin_42712867/article/details/105337052
- 特征选择 https://blog.csdn.net/jiabiao1602/article/details/44975741
- 调参的好例子 https://machinelearningmastery.com/tune-machine-learning-algorithms-in-r/
- Caret set seeds https://stackoverflow.com/questions/13403427/fully-reproducible-parallel-models-using-caret